Creating Molecular Graphs in Graphein#
Graphein provides a set of tools for working with molecular graphs. Under the hood, we wrap RDKit functions for the graph construction. While the main format used is an nx.Graph, each graph, node & edge has its underlying RDKit object stored as metadata so RDKit methods can still be used.
[1]:
# Install Graphein if necessary
# !pip install graphein[extras]
[2]:
import networkx as nx
import logging
logging.getLogger("matplotlib").setLevel(logging.WARNING)
import graphein.molecule as gm
INFO:rdkit:Enabling RDKit 2022.03.1 jupyter extensions
Config#
Similar to Protein Graph Construction, configuration of small molecule graphs is controlled by a `MoleculeGraphConfig <>`__ object.
[3]:
config = gm.MoleculeGraphConfig()
config
[3]:
MoleculeGraphConfig(verbose=False, add_hs=False, edge_construction_functions=[<function add_fully_connected_edges at 0x7fd2d31e5ca0>, <function add_k_nn_edges at 0x7fd2d31e5d30>, <function add_distance_threshold at 0x7fd2d31e5c10>, <function add_atom_bonds at 0x7fd2d31e5790>], node_metadata_functions=[<function atom_type_one_hot at 0x7fd270aa30d0>], edge_metadata_functions=None, graph_metadata_functions=None)
Creating Graphs#
Creating a Molecular Graph from SMILES#
Let’s take a look at a simple molecule, acetylsalicylic acid (or aspirin as it’s more commonly known).
[4]:
graph = gm.construct_graph(smiles="CC(=O)OC1=CC=CC=C1C(=O)O", config=config)
print(graph)
nx.draw(graph)
graph.graph["rdmol"]
CC(=O)OC1=CC=CC=C1C(=O)O
[4]:
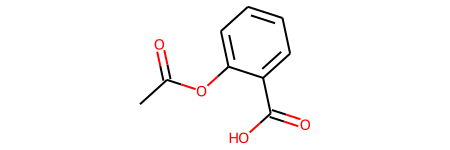
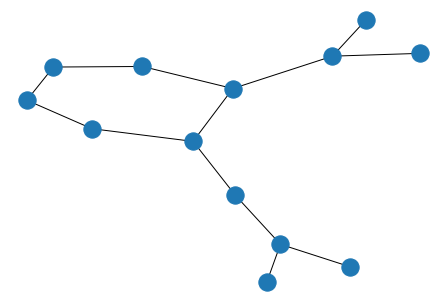
Molecule features are stored as dictionaries on the graph, nodes and edges.
[5]:
# Node metadata
for n, d in graph.nodes(data=True):
print(d)
{'atomic_num': 6, 'element': 'C', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270a5a9a0>, 'coords': None, 'atom_type_one_hot': array([1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0])}
{'atomic_num': 6, 'element': 'C', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270ab1400>, 'coords': None, 'atom_type_one_hot': array([1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0])}
{'atomic_num': 8, 'element': 'O', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270ab1100>, 'coords': None, 'atom_type_one_hot': array([0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0])}
{'atomic_num': 8, 'element': 'O', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270ab10a0>, 'coords': None, 'atom_type_one_hot': array([0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0])}
{'atomic_num': 6, 'element': 'C', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270ab1160>, 'coords': None, 'atom_type_one_hot': array([1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0])}
{'atomic_num': 6, 'element': 'C', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270ab11c0>, 'coords': None, 'atom_type_one_hot': array([1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0])}
{'atomic_num': 6, 'element': 'C', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270ab1220>, 'coords': None, 'atom_type_one_hot': array([1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0])}
{'atomic_num': 6, 'element': 'C', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270ab1280>, 'coords': None, 'atom_type_one_hot': array([1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0])}
{'atomic_num': 6, 'element': 'C', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270ab12e0>, 'coords': None, 'atom_type_one_hot': array([1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0])}
{'atomic_num': 6, 'element': 'C', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270ab1340>, 'coords': None, 'atom_type_one_hot': array([1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0])}
{'atomic_num': 6, 'element': 'C', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270ab13a0>, 'coords': None, 'atom_type_one_hot': array([1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0])}
{'atomic_num': 8, 'element': 'O', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270ab1460>, 'coords': None, 'atom_type_one_hot': array([0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0])}
{'atomic_num': 8, 'element': 'O', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd270ab14c0>, 'coords': None, 'atom_type_one_hot': array([0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0])}
[6]:
# Edge metadata
for u, v, d in graph.edges(data=True):
print(u, v, d)
C:0 C:1 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1ac0>}
C:1 O:2 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1b20>}
C:1 O:3 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1b80>}
O:3 C:4 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1be0>}
C:4 C:5 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1c40>}
C:4 C:9 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1f40>}
C:5 C:6 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1ca0>}
C:6 C:7 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1d00>}
C:7 C:8 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1d60>}
C:8 C:9 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1dc0>}
C:9 C:10 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1e20>}
C:10 O:11 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1e80>}
C:10 O:12 {'kind': {'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270ab1ee0>}
Creating a Molecular Graph from an SDF File#
As SDFs & Mol2 files provide coordinates, we can draw the graph in 3D.
[7]:
import pkg_resources
SDF_PATH = pkg_resources.resource_filename("graphein", "../tests/molecule/test_data/long_test.sdf")
graph = gm.construct_graph(sdf_path=SDF_PATH, config=config)
print(graph)
graph.graph["rdmol"]
long_test
[7]:
[8]:
gm.plotly_molecular_graph(graph)
Creating a Molecular Graph from a Mol2 File#
[9]:
MOL2_PATH = pkg_resources.resource_filename("graphein", "../tests/molecule/test_data/short_test.mol2")
graph = gm.construct_graph(mol2_path=MOL2_PATH, config=config)
print(graph)
graph.graph["rdmol"]
short_test
[9]:
[10]:
gm.plotly_molecular_graph(graph)
Adding Features#
Graphein can add lots of features to graph nodes and edges from RDKit
Node Features#
[11]:
from functools import partial
config = gm.MoleculeGraphConfig(
node_metadata_functions=[
gm.atom_type_one_hot,
gm.atomic_mass,
gm.degree,
gm.total_degree,
gm.total_valence,
gm.explicit_valence,
gm.implicit_valence,
gm.num_explicit_h,
gm.num_implicit_h,
gm.total_num_h,
gm.num_radical_electrons,
gm.formal_charge,
gm.hybridization,
gm.is_aromatic,
gm.is_isotope,
gm.is_ring,
gm.chiral_tag,
partial(gm.is_ring_size, ring_size=5),
partial(gm.is_ring_size, ring_size=7)
]
)
graph = gm.construct_graph(mol2_path=MOL2_PATH, config=config)
for n, d in graph.nodes(data=True):
print(d)
INFO:graphein.molecule.edges.distance:Found: 3 KNN edges
INFO:graphein.molecule.edges.distance:Found: 9 distance edges for radius 5.0
{'atomic_num': 6, 'element': 'C', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd29027ca00>, 'coords': array([ 0.9862, -0.257 , -0.1905]), 'atom_type_one_hot': array([1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]), 'mass': 12.011, 'degree': 1, 'total_degree': 4, 'total_valence': 4, 'explicit_valence': 4, 'implicit_valence': 0, 'num_explicit_h': 3, 'num_implicit_h': 0, 'total_num_h': 3, 'num_radical_electrons': 0, 'formal_charge': 0, 'hybridization': rdkit.Chem.rdchem.HybridizationType.SP3, 'is_aromatic': False, 'is_isotope': 0, 'is_ring': False, 'chiral_tag': rdkit.Chem.rdchem.ChiralType.CHI_UNSPECIFIED, 'is_ring_5': False, 'is_ring_7': False}
{'atomic_num': 6, 'element': 'C', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd29027c9a0>, 'coords': array([-0.2052, 0.2236, 0.6206]), 'atom_type_one_hot': array([1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]), 'mass': 12.011, 'degree': 2, 'total_degree': 4, 'total_valence': 4, 'explicit_valence': 4, 'implicit_valence': 0, 'num_explicit_h': 2, 'num_implicit_h': 0, 'total_num_h': 2, 'num_radical_electrons': 0, 'formal_charge': 0, 'hybridization': rdkit.Chem.rdchem.HybridizationType.SP3, 'is_aromatic': False, 'is_isotope': 0, 'is_ring': False, 'chiral_tag': rdkit.Chem.rdchem.ChiralType.CHI_UNSPECIFIED, 'is_ring_5': False, 'is_ring_7': False}
{'atomic_num': 7, 'element': 'N', 'rdmol_atom': <rdkit.Chem.rdchem.Atom object at 0x7fd29027c8e0>, 'coords': array([-1.4201, 0.2357, -0.1847]), 'atom_type_one_hot': array([0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0]), 'mass': 14.007, 'degree': 1, 'total_degree': 3, 'total_valence': 3, 'explicit_valence': 3, 'implicit_valence': 0, 'num_explicit_h': 2, 'num_implicit_h': 0, 'total_num_h': 2, 'num_radical_electrons': 0, 'formal_charge': 0, 'hybridization': rdkit.Chem.rdchem.HybridizationType.SP3, 'is_aromatic': False, 'is_isotope': 0, 'is_ring': False, 'chiral_tag': rdkit.Chem.rdchem.ChiralType.CHI_UNSPECIFIED, 'is_ring_5': False, 'is_ring_7': False}
Edge Features#
[12]:
config = gm.MoleculeGraphConfig(
edge_metadata_functions=[
gm.add_bond_type,
gm.bond_is_aromatic,
gm.bond_is_conjugated,
gm.bond_is_in_ring,
gm.bond_stereo,
partial(gm.bond_is_in_ring_size, ring_size=5),
partial(gm.bond_is_in_ring_size, ring_size=7)
]
)
graph = gm.construct_graph(mol2_path=MOL2_PATH, config=config)
for u, v, d in graph.edges(data=True):
print(d)
INFO:graphein.molecule.edges.distance:Found: 3 KNN edges
INFO:graphein.molecule.edges.distance:Found: 9 distance edges for radius 5.0
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-C:0
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:1-C:1
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge N:2-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-C:0
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:1-C:1
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge N:2-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-C:0
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:1-C:1
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge N:2-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-C:0
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:1-C:1
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge N:2-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-C:0
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:1-C:1
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge N:2-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-C:0
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:1-C:1
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge N:2-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-C:0
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:0-N:2
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge C:1-C:1
DEBUG:graphein.molecule.features.edges.bonds:No RDKit bond found on edge N:2-N:2
{'kind': {'distance_threshold', 'fully_connected'}, 'bond_type': None, 'aromatic': None, 'conjugated': None, 'ring': None, 'bond_stereo': None, 'ring_size_5': None, 'ring_size_7': None}
{'kind': {'distance_threshold', 'k_nn_1', 'fully_connected', 'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270e09e20>, 'bond_type': rdkit.Chem.rdchem.BondType.SINGLE, '_aromatic': False, 'conjugated': False, 'ring': False, 'bond_stereo': rdkit.Chem.rdchem.BondStereo.STEREONONE, 'ring_size_5': False, 'ring_size_7': False}
{'kind': {'distance_threshold', 'fully_connected'}, 'bond_type': None, 'aromatic': None, 'conjugated': None, 'ring': None, 'bond_stereo': None, 'ring_size_5': None, 'ring_size_7': None}
{'kind': {'distance_threshold', 'fully_connected'}, 'bond_type': None, 'aromatic': None, 'conjugated': None, 'ring': None, 'bond_stereo': None, 'ring_size_5': None, 'ring_size_7': None}
{'kind': {'distance_threshold', 'k_nn_1', 'fully_connected', 'bond'}, 'bond': <rdkit.Chem.rdchem.Bond object at 0x7fd270e090a0>, 'bond_type': rdkit.Chem.rdchem.BondType.SINGLE, '_aromatic': False, 'conjugated': False, 'ring': False, 'bond_stereo': rdkit.Chem.rdchem.BondStereo.STEREONONE, 'ring_size_5': False, 'ring_size_7': False}
{'kind': {'distance_threshold', 'fully_connected'}, 'bond_type': None, 'aromatic': None, 'conjugated': None, 'ring': None, 'bond_stereo': None, 'ring_size_5': None, 'ring_size_7': None}
Graph Features#
Global descriptors of the molecule computed by RDKit can be added to the graph too!
[13]:
config = gm.MoleculeGraphConfig(
graph_metadata_functions=[
gm.mol_descriptors
]
)
graph = gm.construct_graph(mol2_path=MOL2_PATH, config=config)
print(graph.graph)
INFO:graphein.molecule.edges.distance:Found: 3 KNN edges
INFO:graphein.molecule.edges.distance:Found: 9 distance edges for radius 5.0
{'name': 'short_test', 'rdmol': <rdkit.Chem.rdchem.Mol object at 0x7fd280c73dc0>, 'coords': array([[ 0.9862, -0.257 , -0.1905],
[-0.2052, 0.2236, 0.6206],
[-1.4201, 0.2357, -0.1847]]), 'smiles': 'CCN', 'config': MoleculeGraphConfig(verbose=False, add_hs=False, edge_construction_functions=[<function add_fully_connected_edges at 0x7fd2d31e5ca0>, <function add_k_nn_edges at 0x7fd2d31e5d30>, <function add_distance_threshold at 0x7fd2d31e5c10>, <function add_atom_bonds at 0x7fd2d31e5790>], node_metadata_functions=[<function atom_type_one_hot at 0x7fd270aa30d0>], edge_metadata_functions=None, graph_metadata_functions=[<function mol_descriptors at 0x7fd2d31f3280>]), 'MaxEStateIndex': 4.847222222222222, 'MinEStateIndex': 0.75, 'MaxAbsEStateIndex': 4.847222222222222, 'MinAbsEStateIndex': 0.75, 'qed': 0.40623709538988323, 'MolWt': 45.084999999999994, 'HeavyAtomMolWt': 38.028999999999996, 'ExactMolWt': 45.057849223999995, 'NumValenceElectrons': 20, 'NumRadicalElectrons': 0, 'MaxPartialCharge': -0.010576265829206462, 'MinPartialCharge': -0.3307484184390755, 'MaxAbsPartialCharge': 0.3307484184390755, 'MinAbsPartialCharge': 0.010576265829206462, 'FpDensityMorgan1': 2.0, 'FpDensityMorgan2': 2.0, 'FpDensityMorgan3': 2.0, 'BCUT2D_MWHI': 14.489350501685236, 'BCUT2D_MWLOW': 10.840990590774261, 'BCUT2D_CHGHI': 1.3223586846717188, 'BCUT2D_CHGLO': -1.5255036511694584, 'BCUT2D_LOGPHI': 1.1976547781828029, 'BCUT2D_LOGPLOW': -1.871495321468652, 'BCUT2D_MRHI': 3.9983956644101193, 'BCUT2D_MRLOW': 1.1354269715888405, 'BalabanJ': 1.6329931618554523, 'BertzCT': 2.7548875021634682, 'Chi0': 2.7071067811865475, 'Chi0n': 2.284457050376173, 'Chi0v': 2.284457050376173, 'Chi1': 1.4142135623730951, 'Chi1n': 1.1153550716504106, 'Chi1v': 1.1153550716504106, 'Chi2n': 0.408248290463863, 'Chi2v': 0.408248290463863, 'Chi3n': 0.0, 'Chi3v': 0.0, 'Chi4n': 0.0, 'Chi4v': 0.0, 'HallKierAlpha': -0.04, 'Ipc': 2.7548875021634682, 'Kappa1': 2.9600000000000004, 'Kappa2': 1.96, 'Kappa3': 1.9600000000000033, 'LabuteASA': 20.444083839238875, 'PEOE_VSA1': 5.733667477162185, 'PEOE_VSA10': 0.0, 'PEOE_VSA11': 0.0, 'PEOE_VSA12': 0.0, 'PEOE_VSA13': 0.0, 'PEOE_VSA14': 0.0, 'PEOE_VSA2': 0.0, 'PEOE_VSA3': 0.0, 'PEOE_VSA4': 0.0, 'PEOE_VSA5': 0.0, 'PEOE_VSA6': 6.923737199690624, 'PEOE_VSA7': 6.544756405912575, 'PEOE_VSA8': 0.0, 'PEOE_VSA9': 0.0, 'SMR_VSA1': 0.0, 'SMR_VSA10': 0.0, 'SMR_VSA2': 0.0, 'SMR_VSA3': 0.0, 'SMR_VSA4': 5.733667477162185, 'SMR_VSA5': 6.923737199690624, 'SMR_VSA6': 6.544756405912575, 'SMR_VSA7': 0.0, 'SMR_VSA8': 0.0, 'SMR_VSA9': 0.0, 'SlogP_VSA1': 5.733667477162185, 'SlogP_VSA10': 0.0, 'SlogP_VSA11': 0.0, 'SlogP_VSA12': 0.0, 'SlogP_VSA2': 6.544756405912575, 'SlogP_VSA3': 0.0, 'SlogP_VSA4': 0.0, 'SlogP_VSA5': 6.923737199690624, 'SlogP_VSA6': 0.0, 'SlogP_VSA7': 0.0, 'SlogP_VSA8': 0.0, 'SlogP_VSA9': 0.0, 'TPSA': 26.02, 'EState_VSA1': 0.0, 'EState_VSA10': 0.0, 'EState_VSA11': 0.0, 'EState_VSA2': 0.0, 'EState_VSA3': 0.0, 'EState_VSA4': 6.544756405912575, 'EState_VSA5': 0.0, 'EState_VSA6': 0.0, 'EState_VSA7': 6.923737199690624, 'EState_VSA8': 0.0, 'EState_VSA9': 5.733667477162185, 'VSA_EState1': 0.0, 'VSA_EState10': 0.0, 'VSA_EState2': 0.0, 'VSA_EState3': 0.0, 'VSA_EState4': 4.847222222222222, 'VSA_EState5': 0.0, 'VSA_EState6': 0.0, 'VSA_EState7': 0.0, 'VSA_EState8': 2.6527777777777777, 'VSA_EState9': 0.0, 'FractionCSP3': 1.0, 'HeavyAtomCount': 3, 'NHOHCount': 2, 'NOCount': 1, 'NumAliphaticCarbocycles': 0, 'NumAliphaticHeterocycles': 0, 'NumAliphaticRings': 0, 'NumAromaticCarbocycles': 0, 'NumAromaticHeterocycles': 0, 'NumAromaticRings': 0, 'NumHAcceptors': 1, 'NumHDonors': 1, 'NumHeteroatoms': 1, 'NumRotatableBonds': 0, 'NumSaturatedCarbocycles': 0, 'NumSaturatedHeterocycles': 0, 'NumSaturatedRings': 0, 'RingCount': 0, 'MolLogP': -0.03499999999999981, 'MolMR': 14.728400000000002, 'fr_Al_COO': 0, 'fr_Al_OH': 0, 'fr_Al_OH_noTert': 0, 'fr_ArN': 0, 'fr_Ar_COO': 0, 'fr_Ar_N': 0, 'fr_Ar_NH': 0, 'fr_Ar_OH': 0, 'fr_COO': 0, 'fr_COO2': 0, 'fr_C_O': 0, 'fr_C_O_noCOO': 0, 'fr_C_S': 0, 'fr_HOCCN': 0, 'fr_Imine': 0, 'fr_NH0': 0, 'fr_NH1': 0, 'fr_NH2': 1, 'fr_N_O': 0, 'fr_Ndealkylation1': 0, 'fr_Ndealkylation2': 0, 'fr_Nhpyrrole': 0, 'fr_SH': 0, 'fr_aldehyde': 0, 'fr_alkyl_carbamate': 0, 'fr_alkyl_halide': 0, 'fr_allylic_oxid': 0, 'fr_amide': 0, 'fr_amidine': 0, 'fr_aniline': 0, 'fr_aryl_methyl': 0, 'fr_azide': 0, 'fr_azo': 0, 'fr_barbitur': 0, 'fr_benzene': 0, 'fr_benzodiazepine': 0, 'fr_bicyclic': 0, 'fr_diazo': 0, 'fr_dihydropyridine': 0, 'fr_epoxide': 0, 'fr_ester': 0, 'fr_ether': 0, 'fr_furan': 0, 'fr_guanido': 0, 'fr_halogen': 0, 'fr_hdrzine': 0, 'fr_hdrzone': 0, 'fr_imidazole': 0, 'fr_imide': 0, 'fr_isocyan': 0, 'fr_isothiocyan': 0, 'fr_ketone': 0, 'fr_ketone_Topliss': 0, 'fr_lactam': 0, 'fr_lactone': 0, 'fr_methoxy': 0, 'fr_morpholine': 0, 'fr_nitrile': 0, 'fr_nitro': 0, 'fr_nitro_arom': 0, 'fr_nitro_arom_nonortho': 0, 'fr_nitroso': 0, 'fr_oxazole': 0, 'fr_oxime': 0, 'fr_para_hydroxylation': 0, 'fr_phenol': 0, 'fr_phenol_noOrthoHbond': 0, 'fr_phos_acid': 0, 'fr_phos_ester': 0, 'fr_piperdine': 0, 'fr_piperzine': 0, 'fr_priamide': 0, 'fr_prisulfonamd': 0, 'fr_pyridine': 0, 'fr_quatN': 0, 'fr_sulfide': 0, 'fr_sulfonamd': 0, 'fr_sulfone': 0, 'fr_term_acetylene': 0, 'fr_tetrazole': 0, 'fr_thiazole': 0, 'fr_thiocyan': 0, 'fr_thiophene': 0, 'fr_unbrch_alkane': 0, 'fr_urea': 0}