Gene Regulatory Network Tutorial#
In this tutorial we run through GRN graph construction in Graphein. This works by providing a gene_list containing gene names. The interactions between these genes are retrieved from two possible sources: * TRRUST * RegNetwork
This is controlled by providing an edge construction function (add_trrust_edges and add_regnetwork_edges) respectively.
[1]:
import networkx as nx
from functools import partial
from typing import List
import matplotlib.pyplot as plt
import logging
logging.basicConfig(level=logging.INFO)
from easydev import Logging as elogging
elogging.level = "CRITICAL"
from graphein.grn.edges import add_regnetwork_edges, add_trrust_edges
INFO:summarizer.preprocessing.cleaner:'pattern' package not found; tag filters are not available for English
Config Objects#
Global parameters are stored in Config objects. We have a global GRNGraphConfig that contains a TRRUSTConfig and a RegNetworkConfig for parameters relating to each of the two sources.
[2]:
# Initialise Config Object
from graphein.grn.config import GRNGraphConfig
config = GRNGraphConfig()
print(f"GRN Config: \n{config} \n")
print(f"TRRUST Config: \n{config.trrust_config} \n")
print(f"RegNetwork Config: \n{config.regnetwork_config}")
GRN Config:
kwargs={} trrust_config=TRRUSTConfig(filtering_functions=None, root_dir=None, kwargs=None) regnetwork_config=RegNetworkConfig(filtering_functions=None, root_dir=None, kwargs=None)
TRRUST Config:
filtering_functions=None root_dir=None kwargs=None
RegNetwork Config:
filtering_functions=None root_dir=None kwargs=None
Gene List#
This is the list of genes that we wish to construct a GRN graph for
[3]:
gene_list: List[str] = ["AATF", "MYC", "USF1", "SP1", "TP53", "DUSP1"]
Edge Annotation Functions#
These are functions that determine labelling of edges with additional metadata. Here we define one which assigs: * "+" to upregulatory interactions * "-" to downregulatory interactions * "?" to interactons of unknown directionality
Edge Annotation functions take in two nodes (u, v) and the edge data (d)
[4]:
# This function defines the labelling of regulatory directions
def edge_ann_fn(u, v, d):
if "+" in d["regtype"]:
d["regtype"] = "+"
elif "-" in d["regtype"]:
d["regtype"] = "-"
elif "?" in d["regtype"]:
d["regtype"] = "?"
Graph Construction#
[5]:
from graphein.grn.graphs import compute_grn_graph
from graphein.grn.features.node_features import add_sequence_to_nodes
g = compute_grn_graph(
gene_list=gene_list,
edge_construction_funcs=[
partial(add_trrust_edges, trrust_filtering_funcs=config.trrust_config.filtering_functions),
partial(add_regnetwork_edges, regnetwork_filtering_funcs=config.regnetwork_config.filtering_functions),
],
node_annotation_funcs=[add_sequence_to_nodes],#, molecular_weight],
edge_annotation_funcs=[edge_ann_fn],
)
/Users/arianjamasb/opt/anaconda3/envs/graphein-wip/lib/python3.8/site-packages/pandas/core/generic.py:6619: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
return self._update_inplace(result)
WARNING:graphein.grn.parse_regnetwork:RegNetwork is not available. Please check your internet connection or verify at: http://www.regnetworkweb.org
PING regnetworkweb.org (35.171.32.146): 56 data bytes
--- regnetworkweb.org ping statistics ---
1 packets transmitted, 0 packets received, 100.0% packet loss
/Users/arianjamasb/github/graphein/graphein/grn/parse_regnetwork.py:145: DtypeWarning: Columns (1,3) have mixed types.Specify dtype option on import or set low_memory=False.
df = load_RegNetwork_interactions(root_dir)
/Users/arianjamasb/github/graphein/graphein/grn/parse_regnetwork.py:193: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
df["source"] = "RegNetwork"
/Users/arianjamasb/opt/anaconda3/envs/graphein-wip/lib/python3.8/site-packages/pandas/core/generic.py:6619: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
return self._update_inplace(result)
[6]:
for n,d in g.nodes(data=True):
print(n, d)
AATF {'gene_id': 'AATF', 'uniprot_ids': ['Q9NY61'], 'sequence_Q9NY61': 'MAGPQPLALQLEQLLNPRPSEADPEADPEEATAARVIDRFDEGEDGEGDFLVVGSIRKLASASLLDTDKRYCGKTTSRKAWNEDHWEQTLPGSSDEEISDEEGSGDEDSEGLGLEEYDEDDLGAAEEQECGDHRESKKSRSHSAKTPGFSVQSISDFEKFTKGMDDLGSSEEEEDEESGMEEGDDAEDSQGESEEDRAGDRNSEDDGVVMTFSSVKVSEEVEKGRAVKNQIALWDQLLEGRIKLQKALLTTNQLPQPDVFPLFKDKGGPEFSSALKNSHKALKALLRSLVGLQEELLFQYPDTRYLVDGTKPNAGSEEISSEDDELVEEKKQQRRRVPAKRKLEMEDYPSFMAKRFADFTVYRNRTLQKWHDKTKLASGKLGKGFGAFERSILTQIDHILMDKERLLRRTQTKRSVYRVLGKPEPAAQPVPESLPGEPEILPQAPANAHLKDLDEEIFDDDDFYHQLLRELIERKTSSLDPNDQVAMGRQWLAIQKLRSKIHKKVDRKASKGRKLRFHVLSKLLSFMAPIDHTTMNDDARTELYRSLFGQLHPPDEGHGD'}
MYC {'gene_id': 'MYC', 'uniprot_ids': ['P01106'], 'sequence_P01106': 'MPLNVSFTNRNYDLDYDSVQPYFYCDEEENFYQQQQQSELQPPAPSEDIWKKFELLPTPPLSPSRRSGLCSPSYVAVTPFSLRGDNDGGGGSFSTADQLEMVTELLGGDMVNQSFICDPDDETFIKNIIIQDCMWSGFSAAAKLVSEKLASYQAARKDSGSPNPARGHSVCSTSSLYLQDLSAAASECIDPSVVFPYPLNDSSSPKSCASQDSSAFSPSSDSLLSSTESSPQGSPEPLVLHEETPPTTSSDSEEEQEDEEEIDVVSVEKRQAPGKRSESGSPSAGGHSKPPHSPLVLKRCHVSTHQHNYAAPPSTRKDYPAAKRVKLDSVRVLRQISNNRKCTSPRSSDTEENVKRRTHNVLERQRRNELKRSFFALRDQIPELENNEKAPKVVILKKATAYILSVQAEEQKLISEEDLLRKRREQLKHKLEQLRNSCA'}
USF1 {'gene_id': 'USF1', 'uniprot_ids': ['P22415'], 'sequence_P22415': 'MKGQQKTAETEEGTVQIQEGAVATGEDPTSVAIASIQSAATFPDPNVKYVFRTENGGQVMYRVIQVSEGQLDGQTEGTGAISGYPATQSMTQAVIQGAFTSDDAVDTEGTAAETHYTYFPSTAVGDGAGGTTSGSTAAVVTTQGSEALLGQATPPGTGQFFVMMSPQEVLQGGSQRSIAPRTHPYSPKSEAPRTTRDEKRRAQHNEVERRRRDKINNWIVQLSKIIPDCSMESTKSGQSKGGILSKACDYIQELRQSNHRLSEELQGLDQLQLDNDVLRQQVEDLKNKNLLLRAQLRHHGLEVVIKNDSN'}
SP1 {'gene_id': 'SP1', 'uniprot_ids': ['P08047'], 'sequence_P08047': 'MSDQDHSMDEMTAVVKIEKGVGGNNGGNGNGGGAFSQARSSSTGSSSSTGGGGQESQPSPLALLAATCSRIESPNENSNNSQGPSQSGGTGELDLTATQLSQGANGWQIISSSSGATPTSKEQSGSSTNGSNGSESSKNRTVSGGQYVVAAAPNLQNQQVLTGLPGVMPNIQYQVIPQFQTVDGQQLQFAATGAQVQQDGSGQIQIIPGANQQIITNRGSGGNIIAAMPNLLQQAVPLQGLANNVLSGQTQYVTNVPVALNGNITLLPVNSVSAATLTPSSQAVTISSSGSQESGSQPVTSGTTISSASLVSSQASSSSFFTNANSYSTTTTTSNMGIMNFTTSGSSGTNSQGQTPQRVSGLQGSDALNIQQNQTSGGSLQAGQQKEGEQNQQTQQQQILIQPQLVQGGQALQALQAAPLSGQTFTTQAISQETLQNLQLQAVPNSGPIIIRTPTVGPNGQVSWQTLQLQNLQVQNPQAQTITLAPMQGVSLGQTSSSNTTLTPIASAASIPAGTVTVNAAQLSSMPGLQTINLSALGTSGIQVHPIQGLPLAIANAPGDHGAQLGLHGAGGDGIHDDTAGGEEGENSPDAQPQAGRRTRREACTCPYCKDSEGRGSGDPGKKKQHICHIQGCGKVYGKTSHLRAHLRWHTGERPFMCTWSYCGKRFTRSDELQRHKRTHTGEKKFACPECPKRFMRSDHLSKHIKTHQNKKGGPGVALSVGTLPLDSGAGSEGSGTATPSALITTNMVAMEAICPEGIARLANSGINVMQVADLQSINISGNGF'}
TP53 {'gene_id': 'TP53', 'uniprot_ids': ['P04637'], 'sequence_P04637': 'MEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLPSQAMDDLMLSPDDIEQWFTEDPGPDEAPRMPEAAPPVAPAPAAPTPAAPAPAPSWPLSSSVPSQKTYQGSYGFRLGFLHSGTAKSVTCTYSPALNKMFCQLAKTCPVQLWVDSTPPPGTRVRAMAIYKQSQHMTEVVRRCPHHERCSDSDGLAPPQHLIRVEGNLRVEYLDDRNTFRHSVVVPYEPPEVGSDCTTIHYNYMCNSSCMGGMNRRPILTIITLEDSSGNLLGRNSFEVRVCACPGRDRRTEEENLRKKGEPHHELPPGSTKRALPNNTSSSPQPKKKPLDGEYFTLQIRGRERFEMFRELNEALELKDAQAGKEPGGSRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD'}
DUSP1 {'gene_id': 'DUSP1', 'uniprot_ids': ['P28562'], 'sequence_P28562': 'MVMEVGTLDAGGLRALLGERAAQCLLLDCRSFFAFNAGHIAGSVNVRFSTIVRRRAKGAMGLEHIVPNAELRGRLLAGAYHAVVLLDERSAALDGAKRDGTLALAAGALCREARAAQVFFLKGGYEAFSASCPELCSKQSTPMGLSLPLSTSVPDSAESGCSSCSTPLYDQGGPVEILPFLYLGSAYHASRKDMLDALGITALINVSANCPNHFEGHYQYKSIPVEDNHKADISSWFNEAIDFIDSIKNAGGRVFVHCQAGISRSATICLAYLMRTNRVKLDEAFEFVKQRRSIISPNFSFMGQLLQFESQVLAPHCSAEAGSPAMAVLDRGTSTTTVFNFPVSIPVHSTNSALSYLQSPITTSPSC'}
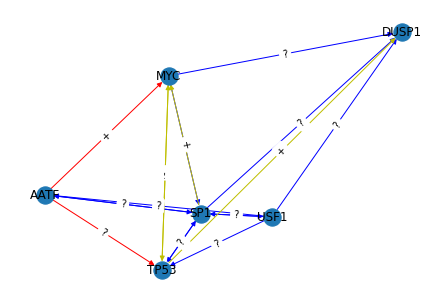
Plotting the Graph#
We now wish to visualise the graph. We can do this with the built in visualisation in NetworkX
[7]:
# Here we define a colouring scheme for the graph
edge_colors = [
"r"
if g[u][v]["kind"] == {"trrust"}
else "b"
if g[u][v]["kind"] == {"regnetwork"}
else "y"
for u, v in g.edges()
]
print(nx.info(g))
pos = nx.spring_layout(g)
nx.draw(g, pos=pos, with_labels=True, edge_color=edge_colors)
edge_labels = {(u, v): g[u][v]["regtype"] for u, v in g.edges}
nx.draw_networkx_edge_labels(g, pos=pos, edge_labels=edge_labels)
plt.show()
Name:
Type: DiGraph
Number of nodes: 6
Number of edges: 22
Average in degree: 3.6667
Average out degree: 3.6667
[8]:
print(g.edges(data=True))
[('AATF', 'TP53', {'kind': {'trrust'}, 'regtype': '?'}), ('AATF', 'MYC', {'kind': {'trrust'}, 'regtype': '+'}), ('AATF', 'SP1', {'kind': {'regnetwork'}, 'regtype': '?'}), ('MYC', 'TP53', {'kind': {'regnetwork', 'trrust'}, 'regtype': '+'}), ('MYC', 'DUSP1', {'kind': {'regnetwork'}, 'regtype': '?'}), ('MYC', 'MYC', {'kind': {'regnetwork'}, 'regtype': '?'}), ('MYC', 'SP1', {'kind': {'regnetwork'}, 'regtype': '?'}), ('USF1', 'AATF', {'kind': {'regnetwork'}, 'regtype': '?'}), ('USF1', 'USF1', {'kind': {'regnetwork'}, 'regtype': '?'}), ('USF1', 'SP1', {'kind': {'regnetwork'}, 'regtype': '?'}), ('USF1', 'TP53', {'kind': {'regnetwork'}, 'regtype': '?'}), ('USF1', 'DUSP1', {'kind': {'regnetwork'}, 'regtype': '?'}), ('SP1', 'MYC', {'kind': {'regnetwork', 'trrust'}, 'regtype': '+'}), ('SP1', 'SP1', {'kind': {'regnetwork'}, 'regtype': '?'}), ('SP1', 'TP53', {'kind': {'regnetwork'}, 'regtype': '?'}), ('SP1', 'AATF', {'kind': {'regnetwork'}, 'regtype': '?'}), ('SP1', 'DUSP1', {'kind': {'regnetwork'}, 'regtype': '?'}), ('SP1', 'USF1', {'kind': {'regnetwork'}, 'regtype': '?'}), ('TP53', 'TP53', {'kind': {'regnetwork', 'trrust'}, 'regtype': '+'}), ('TP53', 'DUSP1', {'kind': {'regnetwork', 'trrust'}, 'regtype': '+'}), ('TP53', 'MYC', {'kind': {'regnetwork', 'trrust'}, 'regtype': '-'}), ('TP53', 'SP1', {'kind': {'regnetwork'}, 'regtype': '?'})]