PPI Networks and Structural Interactomics#
Graphein allows the retrieval of protein-protein interaction data from two widely-used sources: STRING and BioGrid. In conjunction with the structural graph utilities in Graphein, this can enable the development of geometric deep learning methods for structural interactomics.
[1]:
# Install Graphein if necessary
# !pip install graphein
[2]:
import logging
logging.getLogger('urllib3').setLevel(level=logging.CRITICAL)
#logging.getLogger('matplotlib').setLevel(logging.CRITICAL)
Config#
Global parameters are stored in Config objects. We have a global `PPIGraphConfig <https://graphein.ai/modules/graphein.ppi.html#graphein.ppi.config.PPIGraphConfig>`__ that contains a `STRINGConfig <https://graphein.ai/modules/graphein.ppi.html#graphein.ppi.config.STRINGConfig>`__ and a `BioGridConfig <https://graphein.ai/modules/graphein.ppi.html#graphein.ppi.config.BioGridConfig>`__ for parameters relating to each of the two sources.
[3]:
from graphein.ppi.config import PPIGraphConfig
config = PPIGraphConfig()
config
[3]:
PPIGraphConfig(paginate=True, ncbi_taxon_id=9606, kwargs={'STRING_escore': 0.2, 'BIOGRID_throughputTag': 'high'}, string_config=None, biogrid_config=None)
We also need a list of proteins to work from. Let’s use these:
[4]:
protein_list = ["CDC42", "CDK1", "KIF23", "PLK1", "RAC2", "RACGAP1", "RHOA", "RHOB"]
Graph construction is handled with the `compute_ppi_graph() <https://graphein.ai/modules/graphein.ppi.html#graphein.ppi.graphs.compute_ppi_graph>`__ function that takes a list of proteins, a config for the API calls and edge_construction_funcs like `add_string_edges <https://graphein.ai/modules/graphein.ppi.html#graphein.ppi.edges.add_string_edges>`__ and `add_biogrid_edges <https://graphein.ai/modules/graphein.ppi.html#graphein.ppi.edges.add_biogrid_edges>`__.
[5]:
from graphein.ppi.graphs import compute_ppi_graph
from graphein.ppi.edges import add_string_edges, add_biogrid_edges
edge_construction_funcs=[add_string_edges, add_biogrid_edges]
g = compute_ppi_graph(config=config,
protein_list=protein_list,
edge_construction_funcs=edge_construction_funcs
)
DEBUG:graphein.ppi.graphs:Added 8 nodes to graph
DEBUG:graphein.ppi.edges:Added 44 string interaction edges
DEBUG:graphein.ppi.edges:Added 22 biogrid interaction edges
Visualisation#
Let’s see what it looks like! In this plot node colour & size correspond to the degree. The edge colours indicate the source of interactions: * Red - STRING * Blue - BioGrid * Yellow - Both
[6]:
from graphein.ppi.visualisation import plotly_ppi_graph
plotly_ppi_graph(g)
[7]:
g.edges(data=True)
[7]:
EdgeDataView([('CDC42', 'RACGAP1', {'kind': {'string'}}), ('CDC42', 'RHOA', {'kind': {'string', 'biogrid'}}), ('CDC42', 'KIF23', {'kind': {'string'}}), ('CDC42', 'RAC2', {'kind': {'string', 'biogrid'}}), ('CDC42', 'RHOB', {'kind': {'string', 'biogrid'}}), ('CDC42', 'PLK1', {'kind': {'string'}}), ('CDC42', 'CDK1', {'kind': {'string'}}), ('CDK1', 'RACGAP1', {'kind': {'string', 'biogrid'}}), ('CDK1', 'RHOA', {'kind': {'string'}}), ('CDK1', 'KIF23', {'kind': {'string'}}), ('CDK1', 'PLK1', {'kind': {'string'}}), ('KIF23', 'RACGAP1', {'kind': {'string', 'biogrid'}}), ('KIF23', 'PLK1', {'kind': {'string', 'biogrid'}}), ('KIF23', 'RHOA', {'kind': {'string'}}), ('PLK1', 'RACGAP1', {'kind': {'string', 'biogrid'}}), ('PLK1', 'RHOA', {'kind': {'string'}}), ('PLK1', 'RHOB', {'kind': {'biogrid'}}), ('RAC2', 'RHOB', {'kind': {'string'}}), ('RAC2', 'RHOA', {'kind': {'string', 'biogrid'}}), ('RAC2', 'RACGAP1', {'kind': {'string'}}), ('RACGAP1', 'RHOA', {'kind': {'string'}}), ('RACGAP1', 'RHOB', {'kind': {'string', 'biogrid'}}), ('RHOA', 'RHOB', {'kind': {'string', 'biogrid'}})])
Node Metadata#
Cool! Let’s add some metadata to our nodes in the form of sequences and UniProt IDs
[8]:
from graphein.ppi.features.node_features import add_sequence_to_nodes
g = compute_ppi_graph(config=config,
protein_list=protein_list,
edge_construction_funcs= edge_construction_funcs,
node_annotation_funcs=[add_sequence_to_nodes]
)
DEBUG:graphein.ppi.graphs:Added 8 nodes to graph
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING [bioservices:HGNC:119]: URL of the services contains a double //.Check your URL and remove trailing /
WARNING:bioservices:HGNC:URL of the services contains a double //.Check your URL and remove trailing /
DEBUG:graphein.ppi.edges:Added 44 string interaction edges
DEBUG:graphein.ppi.edges:Added 22 biogrid interaction edges
[9]:
for n, d in g.nodes(data=True):
print(d)
{'protein_id': 'CDC42', 'uniprot_ids': ['P60953'], 'sequence_P60953': 'MQTIKCVVVGDGAVGKTCLLISYTTNKFPSEYVPTVFDNYAVTVMIGGEPYTLGLFDTAGQEDYDRLRPLSYPQTDVFLVCFSVVSPSSFENVKEKWVPEITHHCPKTPFLLVGTQIDLRDDPSTIEKLAKNKQKPITPETAEKLARDLKAVKYVECSALTQKGLKNVFDEAILAALEPPEPKKSRRCVLL'}
{'protein_id': 'CDK1', 'uniprot_ids': ['P06493'], 'sequence_P06493': 'MEDYTKIEKIGEGTYGVVYKGRHKTTGQVVAMKKIRLESEEEGVPSTAIREISLLKELRHPNIVSLQDVLMQDSRLYLIFEFLSMDLKKYLDSIPPGQYMDSSLVKSYLYQILQGIVFCHSRRVLHRDLKPQNLLIDDKGTIKLADFGLARAFGIPIRVYTHEVVTLWYRSPEVLLGSARYSTPVDIWSIGTIFAELATKKPLFHGDSEIDQLFRIFRALGTPNNEVWPEVESLQDYKNTFPKWKPGSLASHVKNLDENGLDLLSKMLIYDPAKRISGKMALNHPYFNDLDNQIKKM'}
{'protein_id': 'KIF23', 'uniprot_ids': ['Q02241'], 'sequence_Q02241': 'MKSARAKTPRKPTVKKGSQTNLKDPVGVYCRVRPLGFPDQECCIEVINNTTVQLHTPEGYRLNRNGDYKETQYSFKQVFGTHTTQKELFDVVANPLVNDLIHGKNGLLFTYGVTGSGKTHTMTGSPGEGGLLPRCLDMIFNSIGSFQAKRYVFKSNDRNSMDIQCEVDALLERQKREAMPNPKTSSSKRQVDPEFADMITVQEFCKAEEVDEDSVYGVFVSYIEIYNNYIYDLLEEVPFDPIKPKPPQSKLLREDKNHNMYVAGCTEVEVKSTEEAFEVFWRGQKKRRIANTHLNRESSRSHSVFNIKLVQAPLDADGDNVLQEKEQITISQLSLVDLAGSERTNRTRAEGNRLREAGNINQSLMTLRTCMDVLRENQMYGTNKMVPYRDSKLTHLFKNYFDGEGKVRMIVCVNPKAEDYEENLQVMRFAEVTQEVEVARPVDKAICGLTPGRRYRNQPRGPVGNEPLVTDVVLQSFPPLPSCEILDINDEQTLPRLIEALEKRHNLRQMMIDEFNKQSNAFKALLQEFDNAVLSKENHMQGKLNEKEKMISGQKLEIERLEKKNKTLEYKIEILEKTTTIYEEDKRNLQQELETQNQKLQRQFSDKRRLEARLQGMVTETTMKWEKECERRVAAKQLEMQNKLWVKDEKLKQLKAIVTEPKTEKPERPSRERDREKVTQRSVSPSPVPLSSNYIAQISNGQQLMSQPQLHRRSNSCSSISVASCISEWEQKIPTYNTPLKVTSIARRRQQEPGQSKTCIVSDRRRGMYWTEGREVVPTFRNEIEIEEDHCGRLLFQPDQNAPPIRLRHRRSRSAGDRWVDHKPASNMQTETVMQPHVPHAITVSVANEKALAKCEKYMLTHQELASDGEIETKLIKGDIYKTRGGGQSVQFTDIETLKQESPNGSRKRRSSTVAPAQPDGAESEWTDVETRCSVAVEMRAGSQLGPGYQHHAQPKRKKP'}
{'protein_id': 'PLK1', 'uniprot_ids': ['P53350'], 'sequence_P53350': 'MSAAVTAGKLARAPADPGKAGVPGVAAPGAPAAAPPAKEIPEVLVDPRSRRRYVRGRFLGKGGFAKCFEISDADTKEVFAGKIVPKSLLLKPHQREKMSMEISIHRSLAHQHVVGFHGFFEDNDFVFVVLELCRRRSLLELHKRRKALTEPEARYYLRQIVLGCQYLHRNRVIHRDLKLGNLFLNEDLEVKIGDFGLATKVEYDGERKKTLCGTPNYIAPEVLSKKGHSFEVDVWSIGCIMYTLLVGKPPFETSCLKETYLRIKKNEYSIPKHINPVAASLIQKMLQTDPTARPTINELLNDEFFTSGYIPARLPITCLTIPPRFSIAPSSLDPSNRKPLTVLNKGLENPLPERPREKEEPVVRETGEVVDCHLSDMLQQLHSVNASKPSERGLVRQEEAEDPACIPIFWVSKWVDYSDKYGLGYQLCDNSVGVLFNDSTRLILYNDGDSLQYIERDGTESYLTVSSHPNSLMKKITLLKYFRNYMSEHLLKAGANITPREGDELARLPYLRTWFRTRSAIILHLSNGSVQINFFQDHTKLILCPLMAAVTYIDEKRDFRTYRLSLLEEYGCCKELASRLRYARTMVDKLLSSRSASNRLKAS'}
{'protein_id': 'RAC2', 'uniprot_ids': ['P15153'], 'sequence_P15153': 'MQAIKCVVVGDGAVGKTCLLISYTTNAFPGEYIPTVFDNYSANVMVDSKPVNLGLWDTAGQEDYDRLRPLSYPQTDVFLICFSLVSPASYENVRAKWFPEVRHHCPSTPIILVGTKLDLRDDKDTIEKLKEKKLAPITYPQGLALAKEIDSVKYLECSALTQRGLKTVFDEAIRAVLCPQPTRQQKRACSLL'}
{'protein_id': 'RACGAP1', 'uniprot_ids': ['Q9H0H5'], 'sequence_Q9H0H5': 'MDTMMLNVRNLFEQLVRRVEILSEGNEVQFIQLAKDFEDFRKKWQRTDHELGKYKDLLMKAETERSALDVKLKHARNQVDVEIKRRQRAEADCEKLERQIQLIREMLMCDTSGSIQLSEEQKSALAFLNRGQPSSSNAGNKRLSTIDESGSILSDISFDKTDESLDWDSSLVKTFKLKKREKRRSTSRQFVDGPPGPVKKTRSIGSAVDQGNESIVAKTTVTVPNDGGPIEAVSTIETVPYWTRSRRKTGTLQPWNSDSTLNSRQLEPRTETDSVGTPQSNGGMRLHDFVSKTVIKPESCVPCGKRIKFGKLSLKCRDCRVVSHPECRDRCPLPCIPTLIGTPVKIGEGMLADFVSQTSPMIPSIVVHCVNEIEQRGLTETGLYRISGCDRTVKELKEKFLRVKTVPLLSKVDDIHAICSLLKDFLRNLKEPLLTFRLNRAFMEAAEITDEDNSIAAMYQAVGELPQANRDTLAFLMIHLQRVAQSPHTKMDVANLAKVFGPTIVAHAVPNPDPVTMLQDIKRQPKVVERLLSLPLEYWSQFMMVEQENIDPLHVIENSNAFSTPQTPDIKVSLLGPVTTPEHQLLKTPSSSSLSQRVRSTLTKNTPRFGSKSKSATNLGRQGNFFASPMLK'}
{'protein_id': 'RHOA', 'uniprot_ids': ['P61586'], 'sequence_P61586': 'MAAIRKKLVIVGDGACGKTCLLIVFSKDQFPEVYVPTVFENYVADIEVDGKQVELALWDTAGQEDYDRLRPLSYPDTDVILMCFSIDSPDSLENIPEKWTPEVKHFCPNVPIILVGNKKDLRNDEHTRRELAKMKQEPVKPEEGRDMANRIGAFGYMECSAKTKDGVREVFEMATRAALQARRGKKKSGCLVL'}
{'protein_id': 'RHOB', 'uniprot_ids': ['P62745'], 'sequence_P62745': 'MAAIRKKLVVVGDGACGKTCLLIVFSKDEFPEVYVPTVFENYVADIEVDGKQVELALWDTAGQEDYDRLRPLSYPDTDVILMCFSVDSPDSLENIPEKWVPEVKHFCPNVPIILVANKKDLRSDEHVRTELARMKQEPVRTDDGRAMAVRIQAYDYLECSAKTKEGVREVFETATRAALQKRYGSQNGCINCCKVL'}
Graph Metadata#
Similarly, you can add graph-level metadata from STRING or BioGrid with:
from graphein.ppi.graph_metadata import add_biogrid_metadata, add_string_metadata, add_string_biogrid_metadata
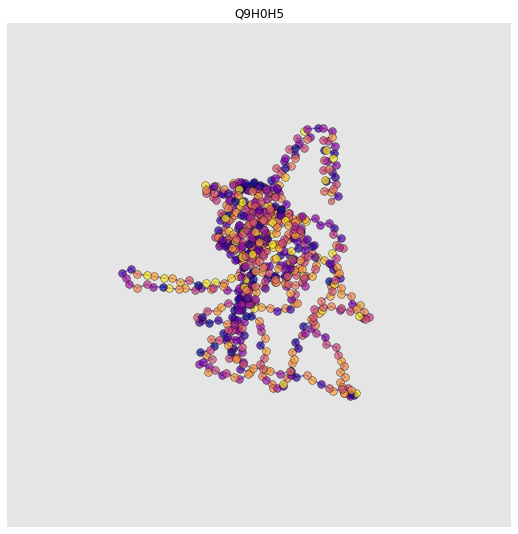
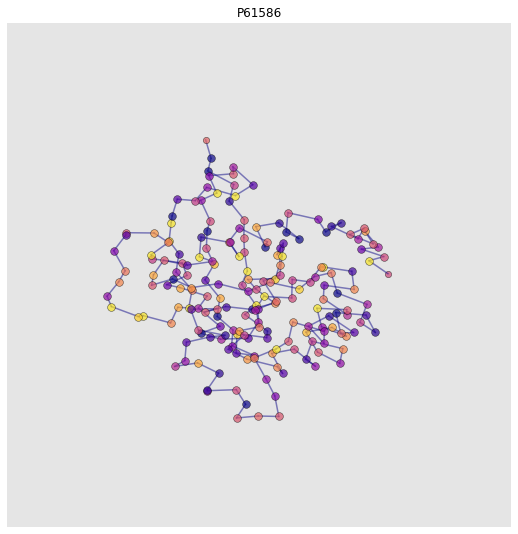
Structural Interactomics with AlphaFold2#
Let’s look at a structural interactome! We can add protein graphs to the nodes in our interaction graph. Cool, eh? We think so
[10]:
import matplotlib.pyplot as plt
logging.getLogger("matplotlib").setLevel(logging.WARNING)
from graphein.protein.config import ProteinGraphConfig
from graphein.protein.graphs import construct_graph
from graphein.protein.utils import download_alphafold_structure
from graphein.protein.visualisation import plot_protein_structure_graph
pg_config = ProteinGraphConfig()
# Iterate over nodes in PPI Graph
for n, d in g.nodes(data=True):
try:
fp = download_alphafold_structure(d['uniprot_ids'][0])[0]
pg = construct_graph(pg_config, pdb_path=fp)
# Add protein graph as node feature
d['protein_graph'] = pg
# Plot
ax = plot_protein_structure_graph(pg, label_node_ids=False, colour_nodes_by="residue_name")
ax.set_title(d["uniprot_ids"][0])
plt.show()
except:
print(f"Failed to construct graph for {d['uniprot_ids'][0]}")
continue
INFO:graphein.protein.utils:Downloaded AlphaFold PDB file for: P60953
-1 / unknown
DEBUG:graphein.protein.graphs:Deprotonating protein. This removes H atoms from the pdb_df dataframe
DEBUG:graphein.protein.graphs:Detected 191 total nodes
DEBUG:graphein.protein.features.nodes.amino_acid:Reading meiler embeddings from: /Users/arianjamasb/github/graphein/graphein/protein/features/nodes/meiler_embeddings.csv
-1 / unknown191
/Users/arianjamasb/github/graphein/graphein/protein/visualisation.py:375: MatplotlibDeprecationWarning:
Axes3D(fig) adding itself to the figure is deprecated since 3.4. Pass the keyword argument auto_add_to_figure=False and use fig.add_axes(ax) to suppress this warning. The default value of auto_add_to_figure will change to False in mpl3.5 and True values will no longer work in 3.6. This is consistent with other Axes classes.

INFO:graphein.protein.utils:Downloaded AlphaFold PDB file for: P06493
-1 / unknown
DEBUG:graphein.protein.graphs:Deprotonating protein. This removes H atoms from the pdb_df dataframe
DEBUG:graphein.protein.graphs:Detected 297 total nodes
-1 / unknown297
/Users/arianjamasb/github/graphein/graphein/protein/visualisation.py:375: MatplotlibDeprecationWarning:
Axes3D(fig) adding itself to the figure is deprecated since 3.4. Pass the keyword argument auto_add_to_figure=False and use fig.add_axes(ax) to suppress this warning. The default value of auto_add_to_figure will change to False in mpl3.5 and True values will no longer work in 3.6. This is consistent with other Axes classes.
-1 / unknown
INFO:graphein.protein.utils:Downloaded AlphaFold PDB file for: Q02241
-1 / unknown
DEBUG:graphein.protein.graphs:Deprotonating protein. This removes H atoms from the pdb_df dataframe
DEBUG:graphein.protein.graphs:Detected 960 total nodes
/Users/arianjamasb/github/graphein/graphein/protein/visualisation.py:375: MatplotlibDeprecationWarning:
Axes3D(fig) adding itself to the figure is deprecated since 3.4. Pass the keyword argument auto_add_to_figure=False and use fig.add_axes(ax) to suppress this warning. The default value of auto_add_to_figure will change to False in mpl3.5 and True values will no longer work in 3.6. This is consistent with other Axes classes.
960
INFO:graphein.protein.utils:Downloaded AlphaFold PDB file for: P53350
-1 / unknown
DEBUG:graphein.protein.graphs:Deprotonating protein. This removes H atoms from the pdb_df dataframe
DEBUG:graphein.protein.graphs:Detected 603 total nodes
-1 / unknown603
/Users/arianjamasb/github/graphein/graphein/protein/visualisation.py:375: MatplotlibDeprecationWarning:
Axes3D(fig) adding itself to the figure is deprecated since 3.4. Pass the keyword argument auto_add_to_figure=False and use fig.add_axes(ax) to suppress this warning. The default value of auto_add_to_figure will change to False in mpl3.5 and True values will no longer work in 3.6. This is consistent with other Axes classes.
INFO:graphein.protein.utils:Downloaded AlphaFold PDB file for: P15153
-1 / unknown
DEBUG:graphein.protein.graphs:Deprotonating protein. This removes H atoms from the pdb_df dataframe
DEBUG:graphein.protein.graphs:Detected 192 total nodes
-1 / unknown192
/Users/arianjamasb/github/graphein/graphein/protein/visualisation.py:375: MatplotlibDeprecationWarning:
Axes3D(fig) adding itself to the figure is deprecated since 3.4. Pass the keyword argument auto_add_to_figure=False and use fig.add_axes(ax) to suppress this warning. The default value of auto_add_to_figure will change to False in mpl3.5 and True values will no longer work in 3.6. This is consistent with other Axes classes.
INFO:graphein.protein.utils:Downloaded AlphaFold PDB file for: Q9H0H5
-1 / unknown
DEBUG:graphein.protein.graphs:Deprotonating protein. This removes H atoms from the pdb_df dataframe
-1 / unknown
DEBUG:graphein.protein.graphs:Detected 632 total nodes
/Users/arianjamasb/github/graphein/graphein/protein/visualisation.py:375: MatplotlibDeprecationWarning:
Axes3D(fig) adding itself to the figure is deprecated since 3.4. Pass the keyword argument auto_add_to_figure=False and use fig.add_axes(ax) to suppress this warning. The default value of auto_add_to_figure will change to False in mpl3.5 and True values will no longer work in 3.6. This is consistent with other Axes classes.
632
INFO:graphein.protein.utils:Downloaded AlphaFold PDB file for: P61586
-1 / unknown
DEBUG:graphein.protein.graphs:Deprotonating protein. This removes H atoms from the pdb_df dataframe
DEBUG:graphein.protein.graphs:Detected 193 total nodes
-1 / unknown193
/Users/arianjamasb/github/graphein/graphein/protein/visualisation.py:375: MatplotlibDeprecationWarning:
Axes3D(fig) adding itself to the figure is deprecated since 3.4. Pass the keyword argument auto_add_to_figure=False and use fig.add_axes(ax) to suppress this warning. The default value of auto_add_to_figure will change to False in mpl3.5 and True values will no longer work in 3.6. This is consistent with other Axes classes.
INFO:graphein.protein.utils:Downloaded AlphaFold PDB file for: P62745
-1 / unknown
DEBUG:graphein.protein.graphs:Deprotonating protein. This removes H atoms from the pdb_df dataframe
DEBUG:graphein.protein.graphs:Detected 196 total nodes
-1 / unknown196
/Users/arianjamasb/github/graphein/graphein/protein/visualisation.py:375: MatplotlibDeprecationWarning:
Axes3D(fig) adding itself to the figure is deprecated since 3.4. Pass the keyword argument auto_add_to_figure=False and use fig.add_axes(ax) to suppress this warning. The default value of auto_add_to_figure will change to False in mpl3.5 and True values will no longer work in 3.6. This is consistent with other Axes classes.

Let’s checkout the metadata again:
[11]:
for n, d in g.nodes(data=True):
print(d)
{'protein_id': 'CDC42', 'uniprot_ids': ['P60953'], 'sequence_P60953': 'MQTIKCVVVGDGAVGKTCLLISYTTNKFPSEYVPTVFDNYAVTVMIGGEPYTLGLFDTAGQEDYDRLRPLSYPQTDVFLVCFSVVSPSSFENVKEKWVPEITHHCPKTPFLLVGTQIDLRDDPSTIEKLAKNKQKPITPETAEKLARDLKAVKYVECSALTQKGLKNVFDEAILAALEPPEPKKSRRCVLL', 'protein_graph': <networkx.classes.graph.Graph object at 0x7f9458b5e0d0>}
{'protein_id': 'CDK1', 'uniprot_ids': ['P06493'], 'sequence_P06493': 'MEDYTKIEKIGEGTYGVVYKGRHKTTGQVVAMKKIRLESEEEGVPSTAIREISLLKELRHPNIVSLQDVLMQDSRLYLIFEFLSMDLKKYLDSIPPGQYMDSSLVKSYLYQILQGIVFCHSRRVLHRDLKPQNLLIDDKGTIKLADFGLARAFGIPIRVYTHEVVTLWYRSPEVLLGSARYSTPVDIWSIGTIFAELATKKPLFHGDSEIDQLFRIFRALGTPNNEVWPEVESLQDYKNTFPKWKPGSLASHVKNLDENGLDLLSKMLIYDPAKRISGKMALNHPYFNDLDNQIKKM', 'protein_graph': <networkx.classes.graph.Graph object at 0x7f9429fb7730>}
{'protein_id': 'KIF23', 'uniprot_ids': ['Q02241'], 'sequence_Q02241': 'MKSARAKTPRKPTVKKGSQTNLKDPVGVYCRVRPLGFPDQECCIEVINNTTVQLHTPEGYRLNRNGDYKETQYSFKQVFGTHTTQKELFDVVANPLVNDLIHGKNGLLFTYGVTGSGKTHTMTGSPGEGGLLPRCLDMIFNSIGSFQAKRYVFKSNDRNSMDIQCEVDALLERQKREAMPNPKTSSSKRQVDPEFADMITVQEFCKAEEVDEDSVYGVFVSYIEIYNNYIYDLLEEVPFDPIKPKPPQSKLLREDKNHNMYVAGCTEVEVKSTEEAFEVFWRGQKKRRIANTHLNRESSRSHSVFNIKLVQAPLDADGDNVLQEKEQITISQLSLVDLAGSERTNRTRAEGNRLREAGNINQSLMTLRTCMDVLRENQMYGTNKMVPYRDSKLTHLFKNYFDGEGKVRMIVCVNPKAEDYEENLQVMRFAEVTQEVEVARPVDKAICGLTPGRRYRNQPRGPVGNEPLVTDVVLQSFPPLPSCEILDINDEQTLPRLIEALEKRHNLRQMMIDEFNKQSNAFKALLQEFDNAVLSKENHMQGKLNEKEKMISGQKLEIERLEKKNKTLEYKIEILEKTTTIYEEDKRNLQQELETQNQKLQRQFSDKRRLEARLQGMVTETTMKWEKECERRVAAKQLEMQNKLWVKDEKLKQLKAIVTEPKTEKPERPSRERDREKVTQRSVSPSPVPLSSNYIAQISNGQQLMSQPQLHRRSNSCSSISVASCISEWEQKIPTYNTPLKVTSIARRRQQEPGQSKTCIVSDRRRGMYWTEGREVVPTFRNEIEIEEDHCGRLLFQPDQNAPPIRLRHRRSRSAGDRWVDHKPASNMQTETVMQPHVPHAITVSVANEKALAKCEKYMLTHQELASDGEIETKLIKGDIYKTRGGGQSVQFTDIETLKQESPNGSRKRRSSTVAPAQPDGAESEWTDVETRCSVAVEMRAGSQLGPGYQHHAQPKRKKP', 'protein_graph': <networkx.classes.graph.Graph object at 0x7f942a2a3cd0>}
{'protein_id': 'PLK1', 'uniprot_ids': ['P53350'], 'sequence_P53350': 'MSAAVTAGKLARAPADPGKAGVPGVAAPGAPAAAPPAKEIPEVLVDPRSRRRYVRGRFLGKGGFAKCFEISDADTKEVFAGKIVPKSLLLKPHQREKMSMEISIHRSLAHQHVVGFHGFFEDNDFVFVVLELCRRRSLLELHKRRKALTEPEARYYLRQIVLGCQYLHRNRVIHRDLKLGNLFLNEDLEVKIGDFGLATKVEYDGERKKTLCGTPNYIAPEVLSKKGHSFEVDVWSIGCIMYTLLVGKPPFETSCLKETYLRIKKNEYSIPKHINPVAASLIQKMLQTDPTARPTINELLNDEFFTSGYIPARLPITCLTIPPRFSIAPSSLDPSNRKPLTVLNKGLENPLPERPREKEEPVVRETGEVVDCHLSDMLQQLHSVNASKPSERGLVRQEEAEDPACIPIFWVSKWVDYSDKYGLGYQLCDNSVGVLFNDSTRLILYNDGDSLQYIERDGTESYLTVSSHPNSLMKKITLLKYFRNYMSEHLLKAGANITPREGDELARLPYLRTWFRTRSAIILHLSNGSVQINFFQDHTKLILCPLMAAVTYIDEKRDFRTYRLSLLEEYGCCKELASRLRYARTMVDKLLSSRSASNRLKAS', 'protein_graph': <networkx.classes.graph.Graph object at 0x7f942aadf220>}
{'protein_id': 'RAC2', 'uniprot_ids': ['P15153'], 'sequence_P15153': 'MQAIKCVVVGDGAVGKTCLLISYTTNAFPGEYIPTVFDNYSANVMVDSKPVNLGLWDTAGQEDYDRLRPLSYPQTDVFLICFSLVSPASYENVRAKWFPEVRHHCPSTPIILVGTKLDLRDDKDTIEKLKEKKLAPITYPQGLALAKEIDSVKYLECSALTQRGLKTVFDEAIRAVLCPQPTRQQKRACSLL', 'protein_graph': <networkx.classes.graph.Graph object at 0x7f9408da3b80>}
{'protein_id': 'RACGAP1', 'uniprot_ids': ['Q9H0H5'], 'sequence_Q9H0H5': 'MDTMMLNVRNLFEQLVRRVEILSEGNEVQFIQLAKDFEDFRKKWQRTDHELGKYKDLLMKAETERSALDVKLKHARNQVDVEIKRRQRAEADCEKLERQIQLIREMLMCDTSGSIQLSEEQKSALAFLNRGQPSSSNAGNKRLSTIDESGSILSDISFDKTDESLDWDSSLVKTFKLKKREKRRSTSRQFVDGPPGPVKKTRSIGSAVDQGNESIVAKTTVTVPNDGGPIEAVSTIETVPYWTRSRRKTGTLQPWNSDSTLNSRQLEPRTETDSVGTPQSNGGMRLHDFVSKTVIKPESCVPCGKRIKFGKLSLKCRDCRVVSHPECRDRCPLPCIPTLIGTPVKIGEGMLADFVSQTSPMIPSIVVHCVNEIEQRGLTETGLYRISGCDRTVKELKEKFLRVKTVPLLSKVDDIHAICSLLKDFLRNLKEPLLTFRLNRAFMEAAEITDEDNSIAAMYQAVGELPQANRDTLAFLMIHLQRVAQSPHTKMDVANLAKVFGPTIVAHAVPNPDPVTMLQDIKRQPKVVERLLSLPLEYWSQFMMVEQENIDPLHVIENSNAFSTPQTPDIKVSLLGPVTTPEHQLLKTPSSSSLSQRVRSTLTKNTPRFGSKSKSATNLGRQGNFFASPMLK', 'protein_graph': <networkx.classes.graph.Graph object at 0x7f9409a859d0>}
{'protein_id': 'RHOA', 'uniprot_ids': ['P61586'], 'sequence_P61586': 'MAAIRKKLVIVGDGACGKTCLLIVFSKDQFPEVYVPTVFENYVADIEVDGKQVELALWDTAGQEDYDRLRPLSYPDTDVILMCFSIDSPDSLENIPEKWTPEVKHFCPNVPIILVGNKKDLRNDEHTRRELAKMKQEPVKPEEGRDMANRIGAFGYMECSAKTKDGVREVFEMATRAALQARRGKKKSGCLVL', 'protein_graph': <networkx.classes.graph.Graph object at 0x7f9448d0b580>}
{'protein_id': 'RHOB', 'uniprot_ids': ['P62745'], 'sequence_P62745': 'MAAIRKKLVVVGDGACGKTCLLIVFSKDEFPEVYVPTVFENYVADIEVDGKQVELALWDTAGQEDYDRLRPLSYPDTDVILMCFSVDSPDSLENIPEKWVPEVKHFCPNVPIILVANKKDLRSDEHVRTELARMKQEPVRTDDGRAMAVRIQAYDYLECSAKTKEGVREVFETATRAALQKRYGSQNGCINCCKVL', 'protein_graph': <networkx.classes.graph.Graph object at 0x7f9408c028e0>}
Plot SARS-CoV-2 host-virus proteins (Gordon et al., 2020)#
List of human interacting proteins (protein_list) retrieved from:
Gordon et al. (2020). A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. [paper]
[12]:
protein_list = ['AP3B1', 'BRD4', 'BRD2', 'CWC27', 'ZC3H18', 'SLC44A2', 'PMPCB',
'YIF1A', 'ATP1B1', 'ACADM', 'ETFA', 'STOM', 'GGCX', 'ATP6V1A',
'PSMD8', 'REEP5', 'PMPCA', 'ANO6', 'PITRM1', 'SLC30A9', 'FASTKD5',
'SLC30A7', 'TUBGCP3', 'COQ8B', 'SAAL1', 'REEP6', 'INTS4',
'SLC25A21', 'TUBGCP2', 'TARS2', 'RTN4', 'FAM8A1', 'AASS', 'AKAP8L',
'AAR2', 'BZW2', 'RRP9', 'PABPC1', 'CSNK2A2', 'CSNK2B', 'G3BP1',
'PABPC4', 'LARP1', 'FAM98A', 'SNIP1', 'UPF1', 'MOV10', 'G3BP2',
'DDX21', 'RBM28', 'RPL36', 'GOLGA7', 'ZDHHC5', 'POLA1', 'PRIM1',
'PRIM2', 'POLA2', 'COLGALT1', 'PKP2', 'AP2A2', 'GFER', 'ERGIC1',
'AP2M1', 'GRPEL1', 'TBCA', 'SBNO1', 'BCKDK', 'AKAP8', 'MYCBP2',
'SLU7', 'RIPK1', 'UBAP2L', 'TYSND1', 'PDZD11', 'PRRC2B', 'UBAP2',
'ZNF318', 'CRTC3', 'USP54', 'ZC3H7A', 'LARP4B', 'RBM41', 'TCF12',
'PPIL3', 'PLEKHA5', 'TBKBP1', 'CIT', 'HSBP1', 'PCNT', 'CEP43',
'PRKAR2A', 'PRKACA', 'PRKAR2B', 'RDX', 'CENPF', 'TLE1', 'TLE3',
'TLE5', 'GOLGA3', 'GOLGA2', 'GOLGB1', 'GRIPAP1', 'CEP350',
'PDE4DIP', 'CEP135', 'CEP68', 'CNTRL', 'ERC1', 'GCC2', 'CLIP4',
'NIN', 'CEP112', 'MIPOL1', 'USP13', 'GCC1', 'JAKMIP1', 'CDK5RAP2',
'AKAP9', 'GORASP1', 'FYCO1', 'C1orf50', 'CEP250', 'TBK1', 'HOOK1',
'NINL', 'GLA', 'IMPDH2', 'SIRT5', 'NUTF2', 'ARF6', 'RNF41',
'SLC27A2', 'EIF4E2', 'POR', 'RAP1GDS1', 'WASHC4', 'FKBP15',
'GIGYF2', 'IDE', 'TIMM10', 'ALG11', 'NUP210', 'TIMM29', 'DNAJC11',
'TIMM10B', 'TIMM9', 'HDAC2', 'GPX1', 'TRMT1', 'ATP5MG', 'ATP6AP1',
'SIGMAR1', 'ATP13A3', 'AGPS', 'CYB5B', 'ACSL3', 'CYB5R3', 'RALA',
'COMT', 'RAB5C', 'RAB7A', 'RAB8A', 'RAB2A', 'RAB10', 'RAB14',
'RHOA', 'RAB1A', 'GNB1', 'GNG5', 'LMAN2', 'MOGS', 'TOR1AIP1',
'MTARC1', 'QSOX2', 'HS2ST1', 'NDUFAF2', 'SCCPDH', 'SCARB1',
'NAT14', 'DCAKD', 'FAM162A', 'DNAJC19', 'SELENOS', 'PTGES2',
'RAB18', 'MPHOSPH10', 'SRP72', 'ATE1', 'NSD2', 'SRP19', 'SRP54',
'MRPS25', 'DDX10', 'LARP7', 'MEPCE', 'NGDN', 'EXOSC8', 'NARS2',
'NOL10', 'CCDC86', 'SEPSECS', 'EXOSC5', 'EXOSC3', 'AATF', 'HECTD1',
'MRPS2', 'MRPS5', 'EXOSC2', 'MRPS27', 'GTF2F2', 'FBN1', 'FBN2',
'NUP214', 'NUP62', 'DCAF7', 'EIF4H', 'NUP54', 'MIB1', 'SPART',
'NEK9', 'ZNF503', 'NUP88', 'NUP58', 'MAT2B', 'FBLN5', 'PPT1',
'CUL2', 'MAP7D1', 'THTPA', 'ZYG11B', 'TIMM8B', 'RBX1', 'ELOC',
'ELOB', 'HMOX1', 'TRIM59', 'ARL6IP6', 'VPS39', 'CLCC1', 'VPS11',
'SUN2', 'ALG5', 'STOML2', 'NUP98', 'RAE1', 'MTCH1', 'HEATR3',
'MDN1', 'PLOD2', 'TOR1A', 'STC2', 'PLAT', 'ITGB1', 'CISD3',
'COL6A1', 'PVR', 'DNMT1', 'LOX', 'PCSK6', 'INHBE', 'NPC2', 'MFGE8',
'OS9', 'NPTX1', 'POGLUT2', 'POGLUT3', 'ERO1B', 'PLD3', 'FOXRED2',
'CHPF', 'PUSL1', 'EMC1', 'GGH', 'ERLEC1', 'IL17RA', 'NGLY1',
'HS6ST2', 'SDF2', 'NEU1', 'GDF15', 'TM2D3', 'ERP44', 'EDEM3',
'SIL1', 'POFUT1', 'SMOC1', 'PLEKHF2', 'FBXL12', 'UGGT2', 'CHPF2',
'ADAMTS1', 'HYOU1', 'FKBP7', 'ADAM9', 'FKBP10', 'SLC9A3R1',
'CHMP2A', 'CSDE1', 'TOMM70', 'MARK3', 'MARK2', 'DPH5', 'DCTPP1',
'MARK1', 'PTBP2', 'BAG5', 'UBXN8', 'GPAA1', 'WFS1', 'ABCC1',
'F2RL1', 'SCAP', 'DPY19L1', 'TMEM97', 'SLC30A6', 'TAPT1', 'ERMP1',
'NLRX1', 'RETREG3', 'PIGO', 'FAR2', 'ECSIT', 'ALG8', 'TMEM39B',
'GHITM', 'ACAD9', 'NDFIP2', 'BCS1L', 'NDUFAF1', 'TMED5', 'NDUFB9',
'PIGS']
[13]:
import networkx as nx
config = PPIGraphConfig()
g = compute_ppi_graph(config=config, protein_list=protein_list, edge_construction_funcs=edge_construction_funcs)
plotly_ppi_graph(g, node_size_multiplier=1, width=1000, height=1000, layout=nx.layout.circular_layout, edge_opacity=0.2)
DEBUG:graphein.ppi.graphs:Added 332 nodes to graph
DEBUG:graphein.ppi.edges:Added 1856 string interaction edges
DEBUG:graphein.ppi.edges:Added 1795 biogrid interaction edges